|
I am a Research Scientist at NVIDIA, working on AI for Genomics. I got my Ph.D. in the Computer Science at Northwestern University in 2025, advised by Prof. Han Liu. I received my Master in Computer Science at Northwestern University and Bachelor in Mathematics at Zhejiang University. I did several research interns at NVIDIA, Amazon, and Snapchat. Email / CV / Google Scholar / Linkedin / Github |

|
|
My research focuses on Large Language Models (LLM) and Deep Learning (DL). I mainly work on LLM for Genomics. I have also worked several projects on conversational AI and user modeling. |
|
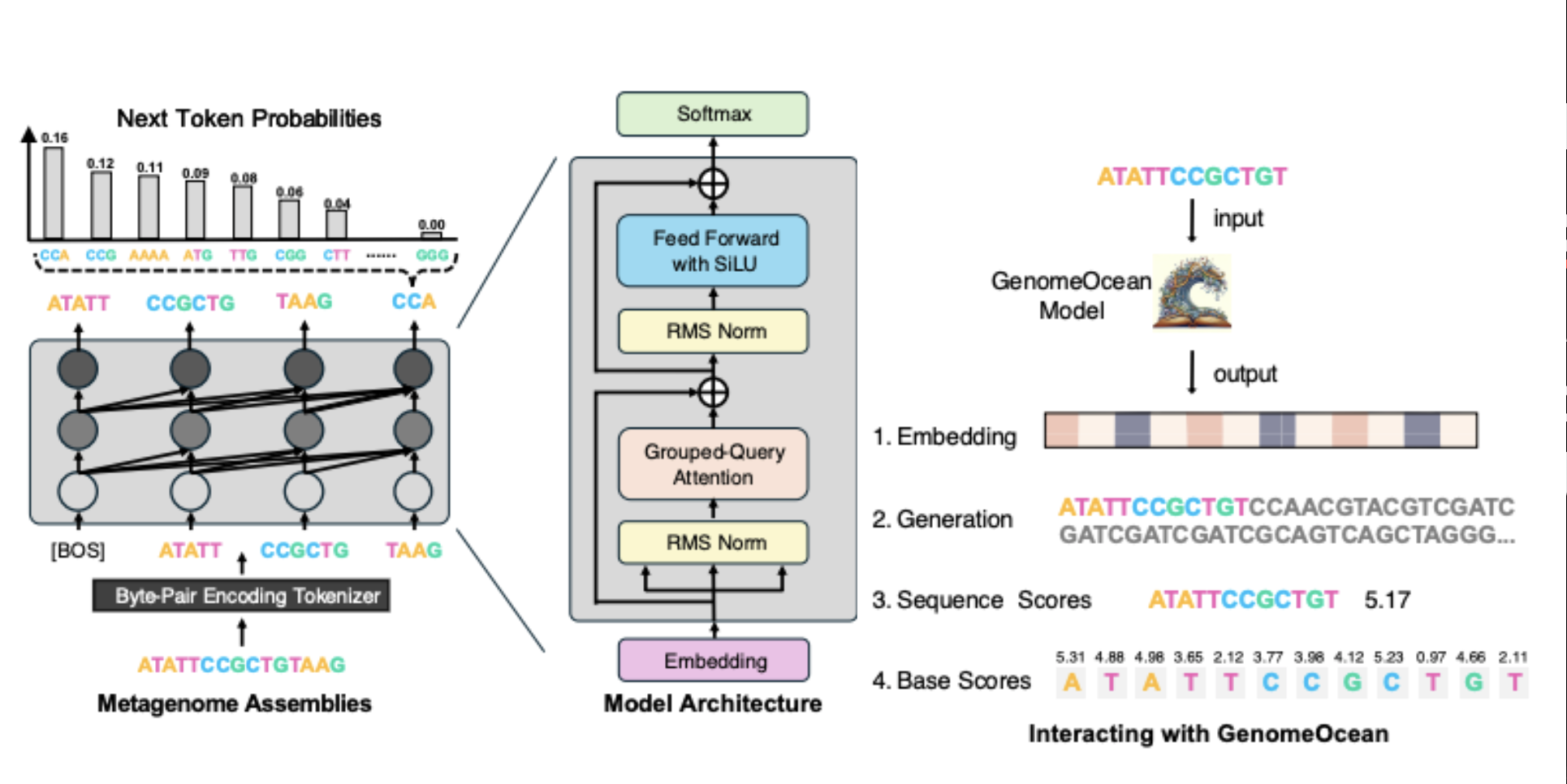
Zhihan Zhou, Robert Riley, Satria Kautsar, Weimin Wu, Rob Egan, Steven Hofmeyr, Shira Goldhaber-Gordon, Mutian Yu, Harrison Ho, Fengchen Liu, Feng Chen, Rachael Morgan-Kiss, Lizhen Shi, Han Liu, Zhong Wang Preprint, 2025 paper / code / model GenomeOcean is a 4B generative genome foundation model which produces protein-coding genes that respect evolutionary constraints and synthesizes large genomic modules such as biosynthetic gene clusters. |
|
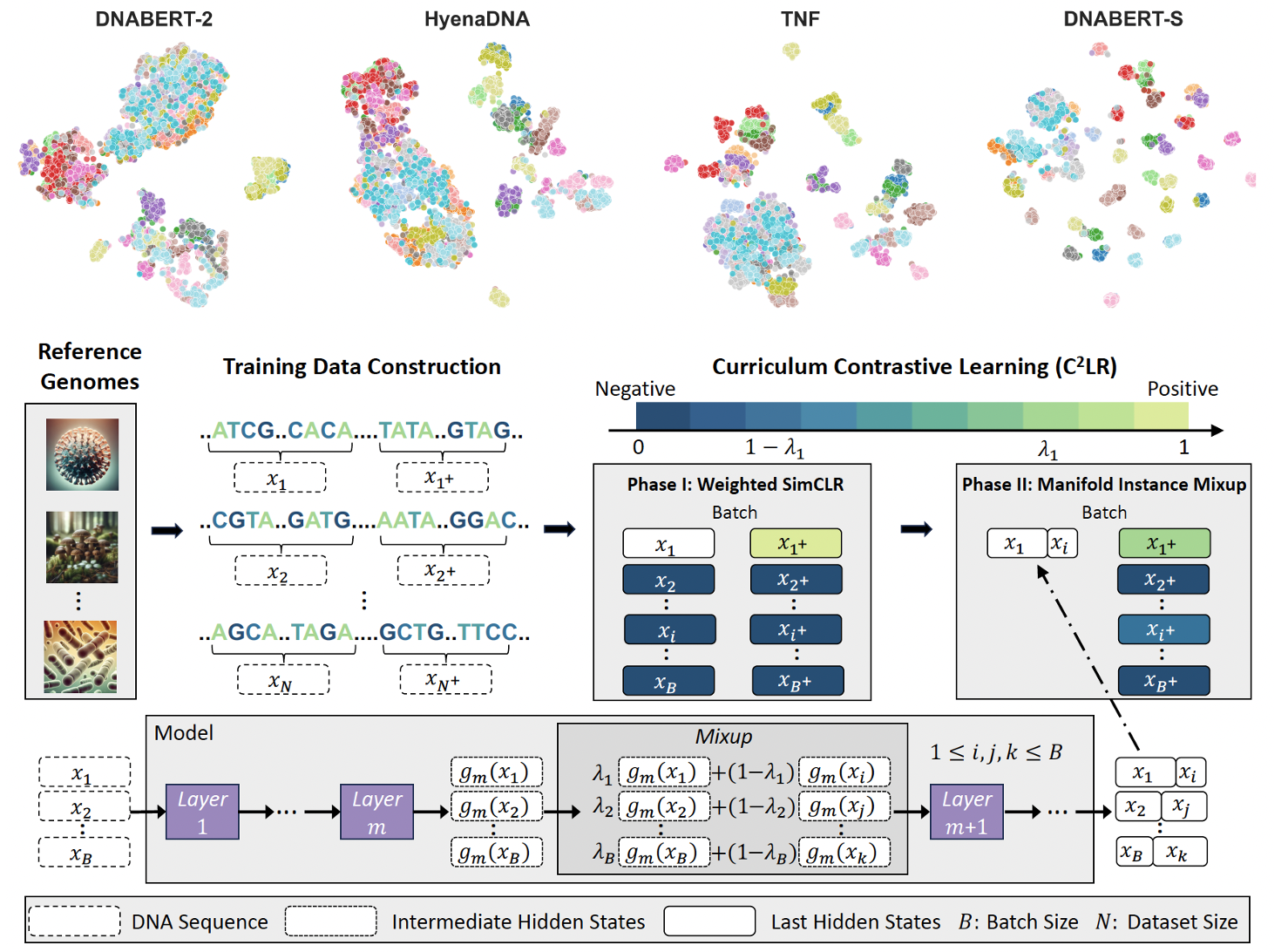
Zhihan Zhou*, Weimin Wu*, Harrison Ho, Jiayi Wang, Lizhen Shi, Ramana V Davuluri, Zhong Wang, Han Liu (*: co-first author) ISMB 2025 paper / code / model DNABERT-S is a foundation model based on DNABERT-2 specifically designed for generating DNA embedding that naturally clusters and segregates genome of different species in the embedding space. |
|
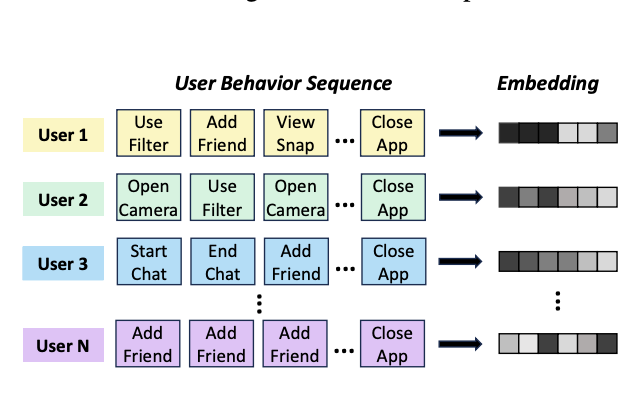
Zhihan Zhou*, Qixiang Fang*, Leonardo Neves, Yozen Liu, Francesco Barbieri, Han Liu, Maarten W. Bos, Ron Dotsch (*: co-first author) Preprint, 2024 paper Seamlessly integrating past and present user behavior sequences for user embedding. |
|
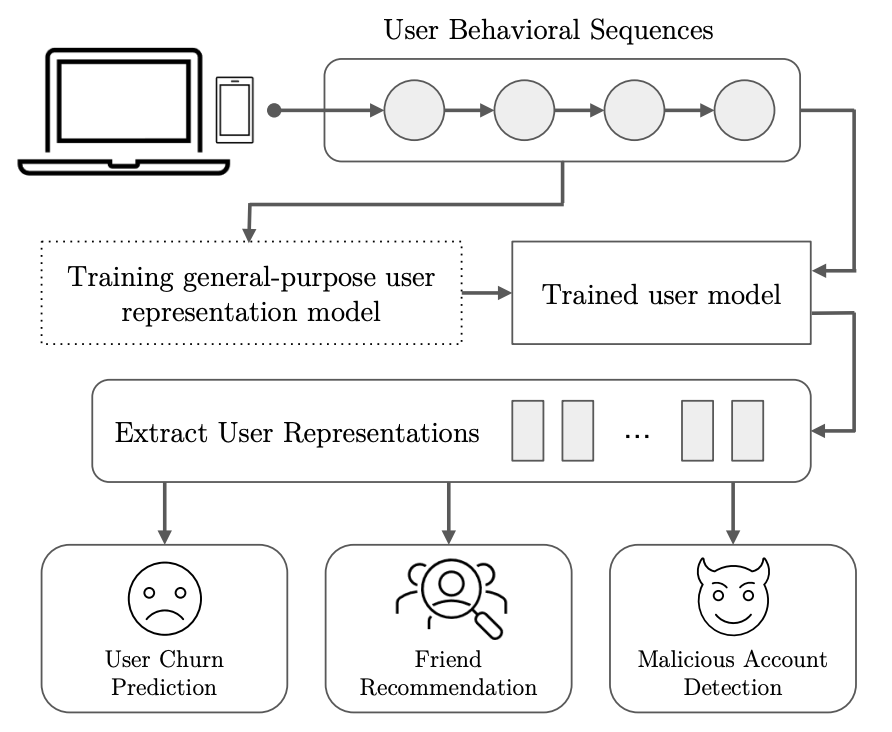
Qixiang Fang*, Zhihan Zhou*, Francesco Barbieri, Yozen Liu, Leonardo Neves, Dong Nguyen, Daniel L. Oberski, Maarten W. Bos, Ron Dotsch (*: co-first author) SIGIR 2024 paper A Transformer-based model that learns general-purpose user embedding from SnapChat user behavioral logs. |
|
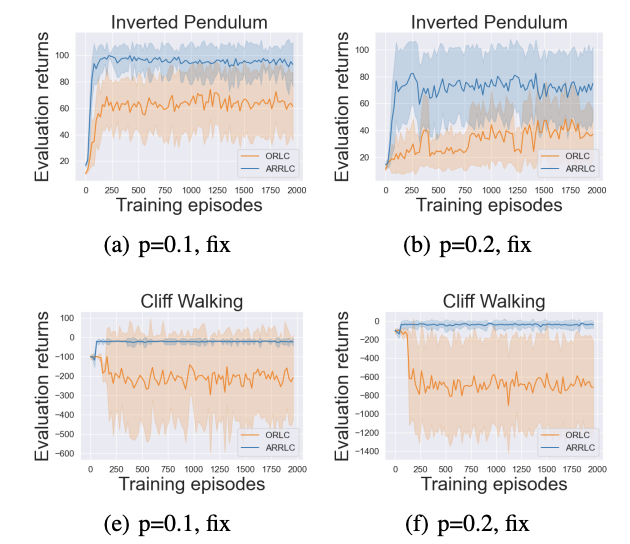
Guanlin Liu, Zhihan Zhou, Han Liu, Lifeng Lai Preprint, 2023 paper A novel approach for solving action robust RL problems with probabilistic policy execution uncertainty. |
|
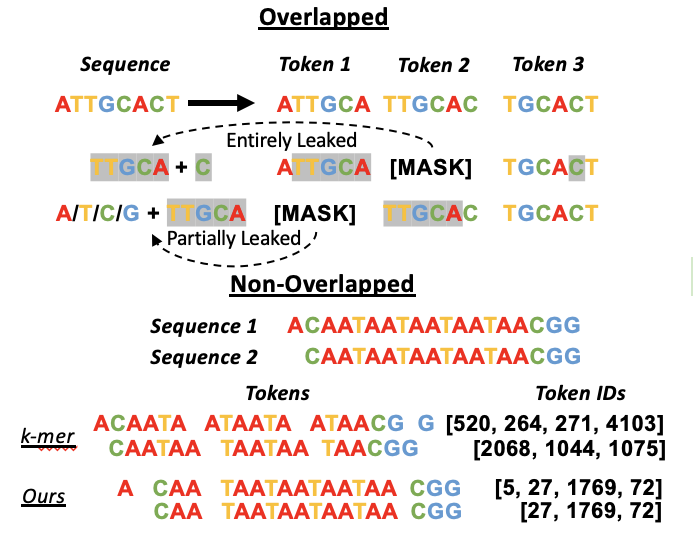
Zhihan Zhou, Yanrong Ji, Weijian Li, Pratik Dutta, Ramana Davuluri, Han Liu ICLR 2024 paper / code / model We introduce DNABERT-2, an efficient and effective foundation model for multi-species genome that achieves state-of-the-art performance with 20 time less parameters. We also provide a benchmark Genome Understanding Evaluation (GUE) containing 28 datasets across 7 tasks. |
|
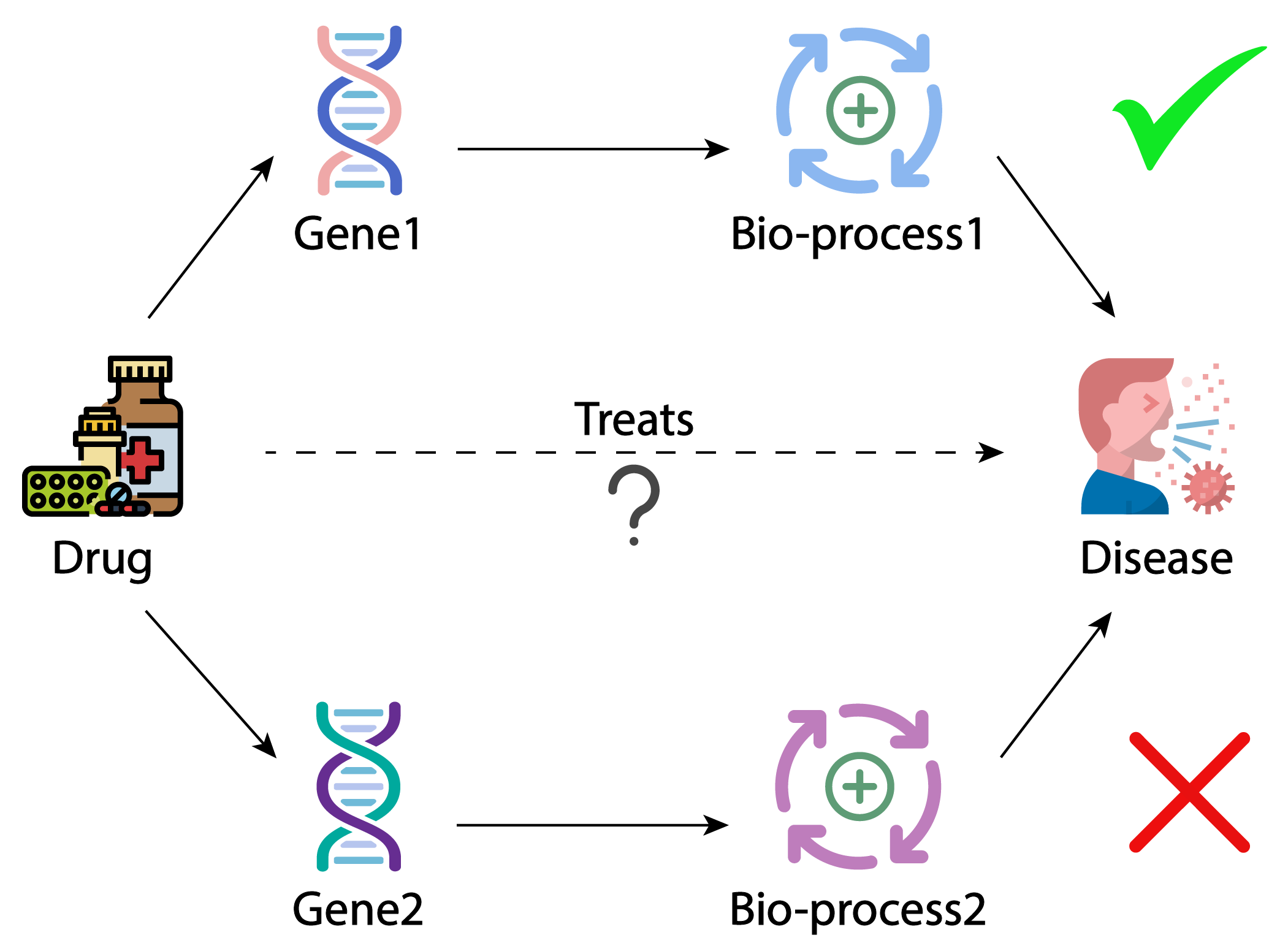
Chunyu Ma, Zhihan Zhou, Han Liu, David Koslicki GigaScience, 2023 code Optimizing Graph Neural Networks and Reinforcement Learning for link prediction and path finding on one of the largest biomedical knowledge graphs. |
|
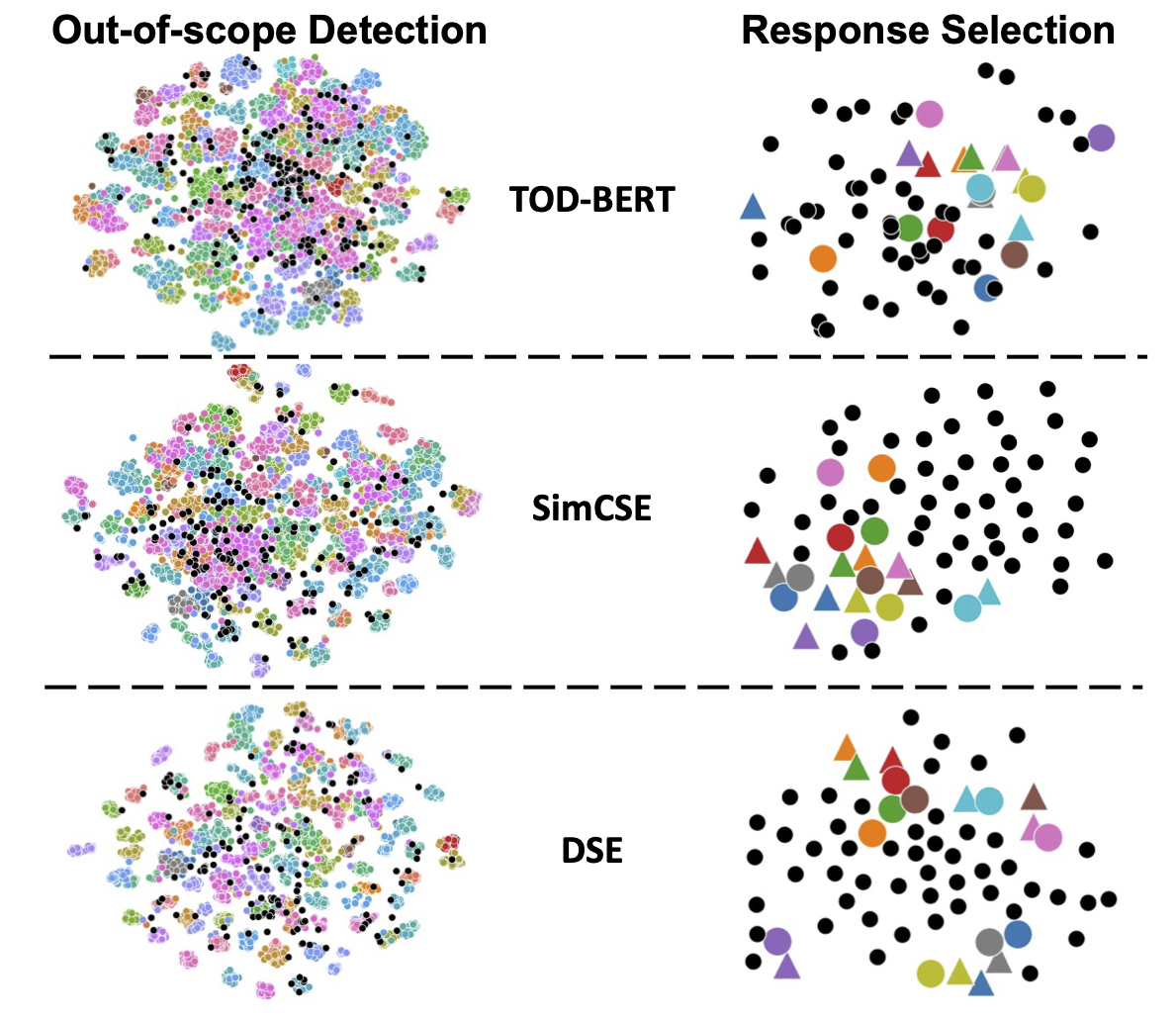
Zhihan Zhou, Dejiao Zhang, Wei Xiao, Nicholas Dingwall, Xiaofei Ma, Andrew O Arnold, Bing Xiang NAACL 2022 paper / code Introduce DSE, a pre-trained language model that achieves great performance in few-shot dialogue understanding. Trained by contrastive learning on consecutive utterances of dialogues. |
|
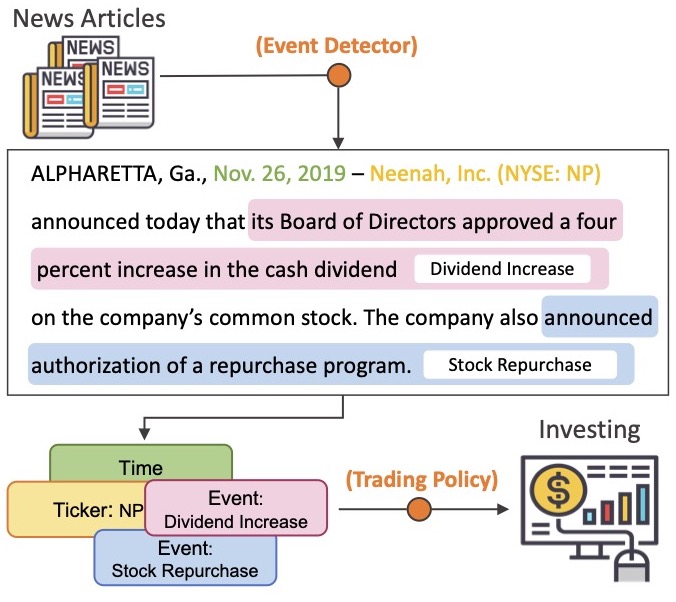
Zhihan Zhou, Liqian Ma, Han Liu ACL 2021, Findings paper / video / code Analyze the impact of news articles on the stock market. Consider corporate events are the driven force and introduce methods for corporate event detection. Provide new datasets for text-based stock prediction and event detection. |
|
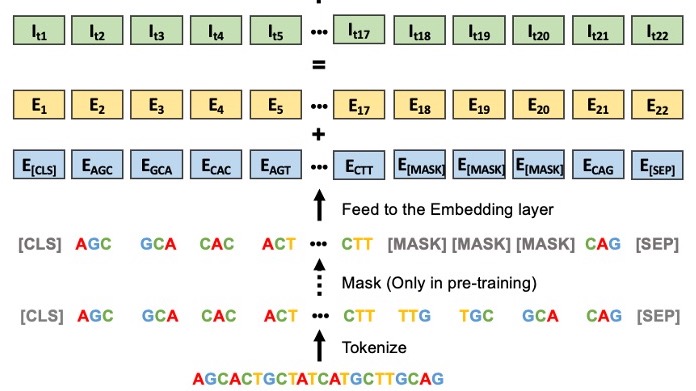
Yanrong Ji*, Zhihan Zhou*, Han Liu, Ramana V Davuluri (*: co-first author) Bioinformatics, 2021 paper / code / model Introduce new paradigm for DNA analysis. Pre-trained on human genome and achieves state-of-the-art performance in a wide range of DNA analysis problems including promoter prediction and splice prediction. |
|
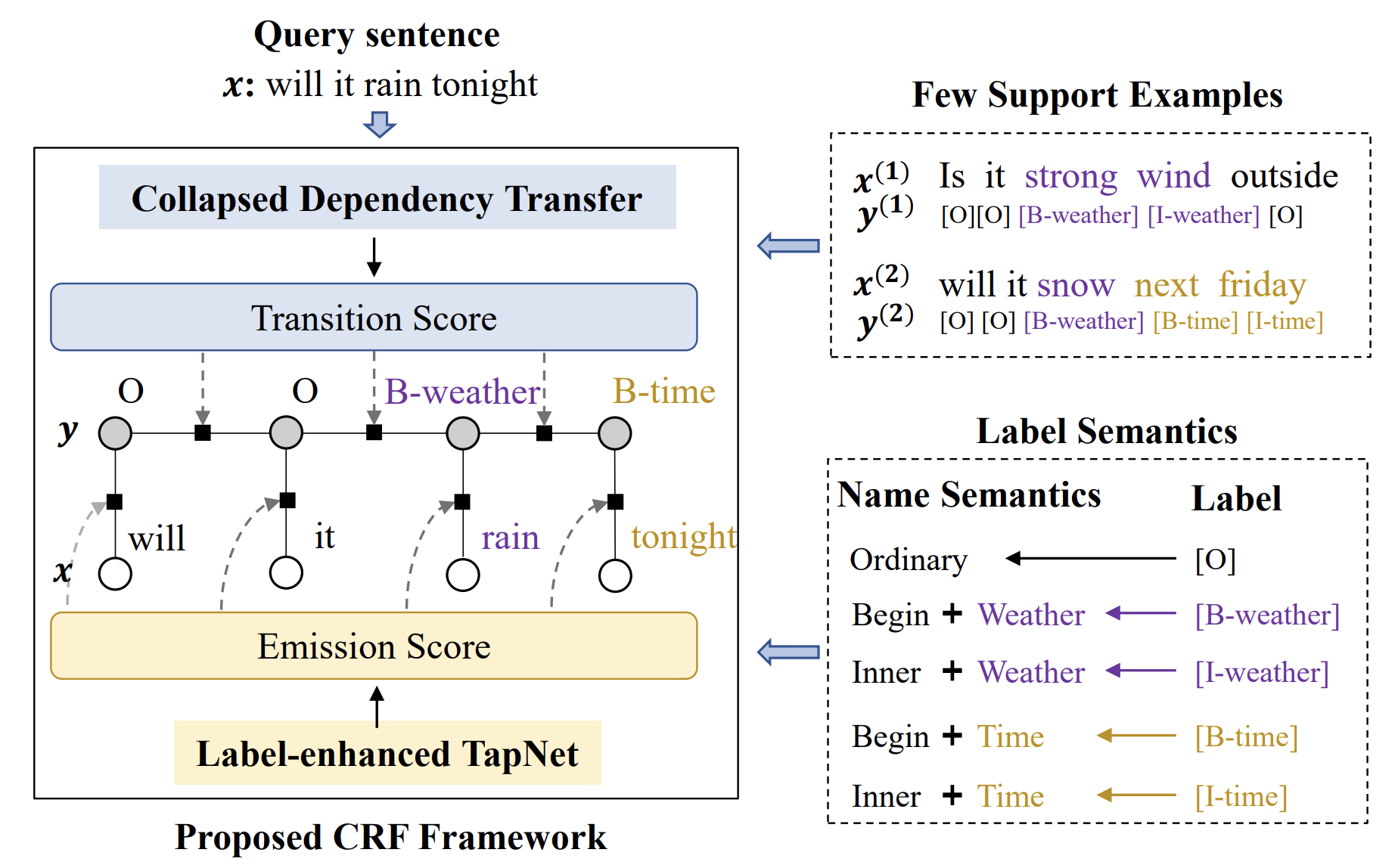
Yutai Hou, Wanxiang Che, Yongkui Lai, Zhihan Zhou, Yijia Liu, Han Liu, Ting Liu ACL 2020 paper / code Introduce novel method for few-shot sequence labeling by incorporating label information and learned label transfer into few-shot prediction. |
|
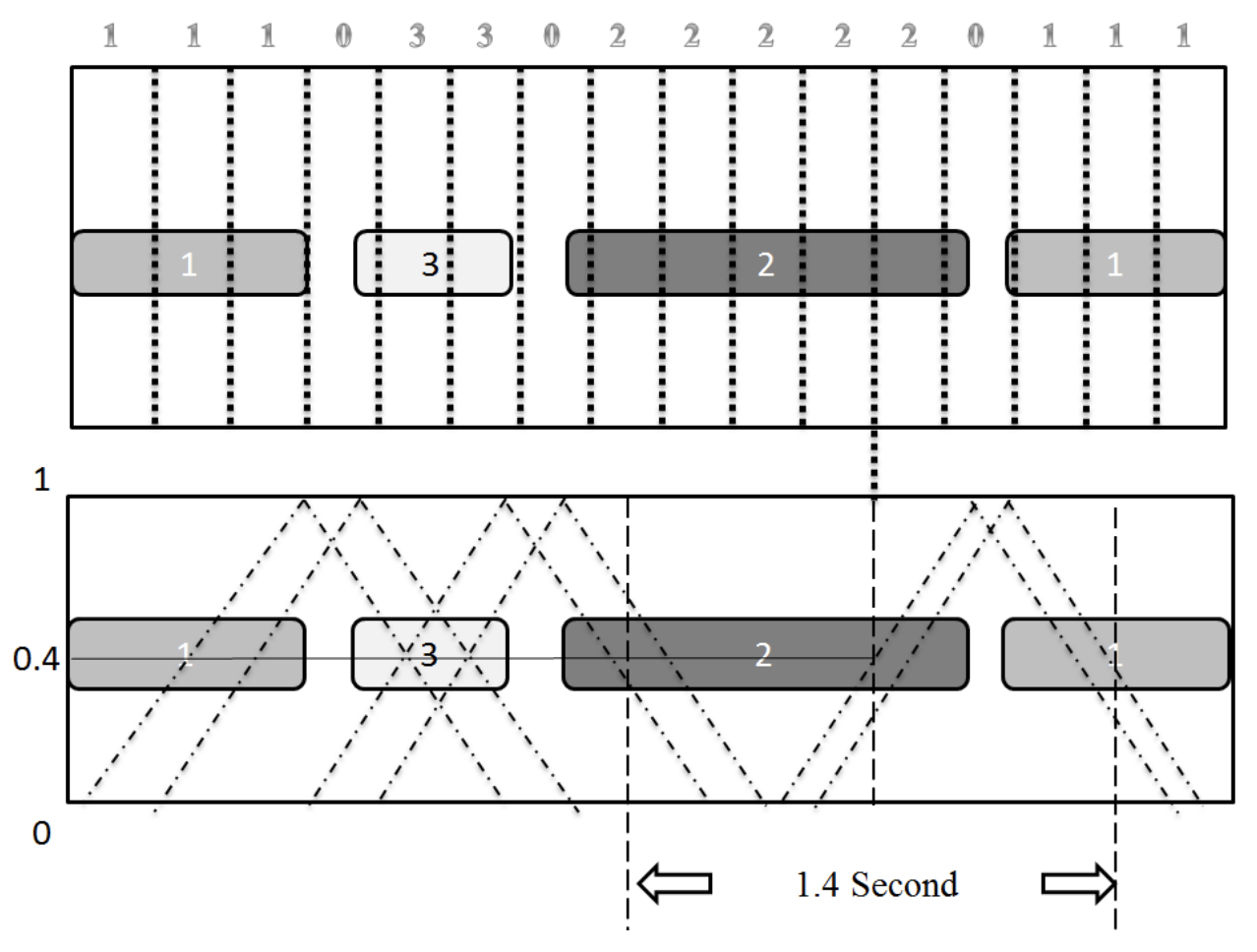
Zhihan Zhou, Yichi Zhang, Zhiyao Duan ICASSP 2018 paper Combines speaker identity predictions and speaker change rate predictions for joint speaker diarization and recognition. |
|
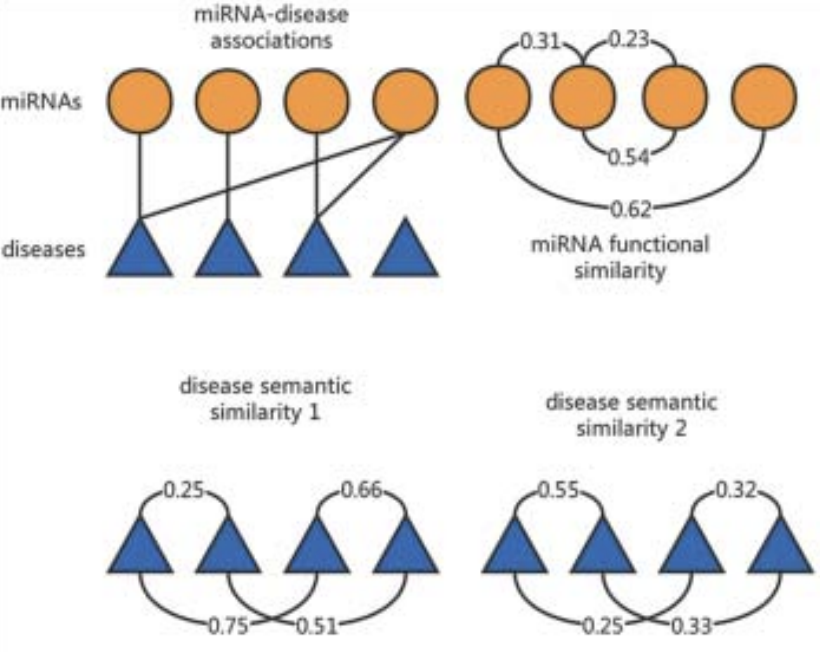
Xing Chen*, Zhihan Zhou*, Yan Zhao (*: co-first author) RNA Biology, 2018 paper Perform ensemble learning on multiple graph learning method to achieve accurate miRNA-disease association prediction. |
|
|

|
Reviewer Serve as reviewer for NeurIPS, ICLR, ICML, ACL, EMNLP, NAACL, AISTATS, etc. Teaching Assistant COMP_SCI 348 Introduction to Artificial Intelligence COMP_SCI 397 Seminar in Statistical Language Modeling COMP_SCI 496-6 Advanced Topics on Deep Learning COMP_SCI 496-9 Statistical Machine learning COMP_SCI 497 Deep Learning for Natural Language Processing STATS 359 Data Science Project |
|
This website is built upon link. Thank Jon Barron for releasing source code and Qinjie for helping.
|